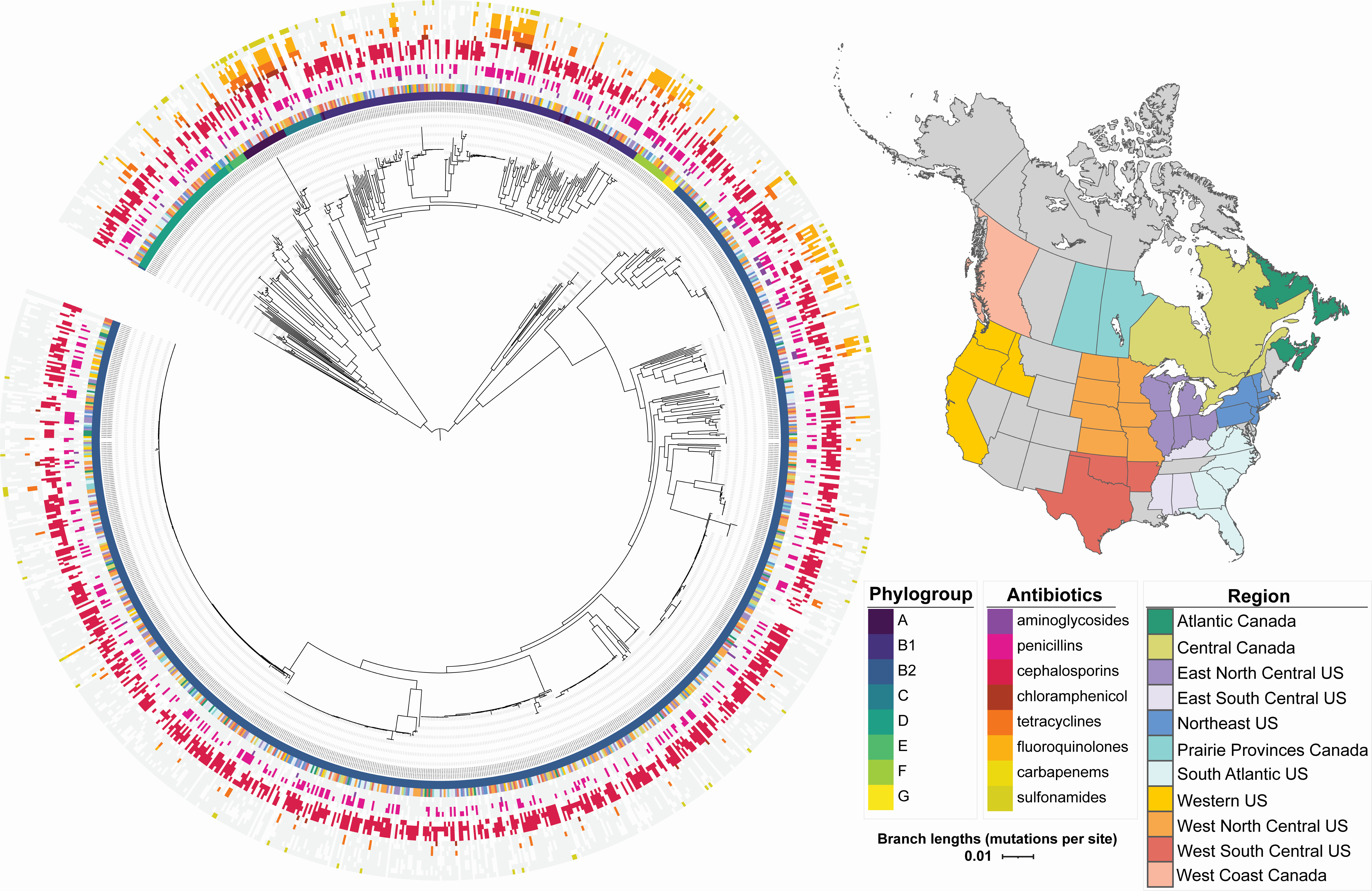
E. coli AMR in dogs
We investigated antimicrobial resistance phenotypes in E. coli samples taken from dogs, and discovered a new potential mechanism of resistance.
I am a 2024 graduate of Cornell's combined DVM-PhD program, and I am broadly interested in wildlife health. I have experience in molecular epidemiology, pathogen evolution and conservation genetics. A full list of my publications can be found on Google Scholar.

We investigated antimicrobial resistance phenotypes in E. coli samples taken from dogs, and discovered a new potential mechanism of resistance.
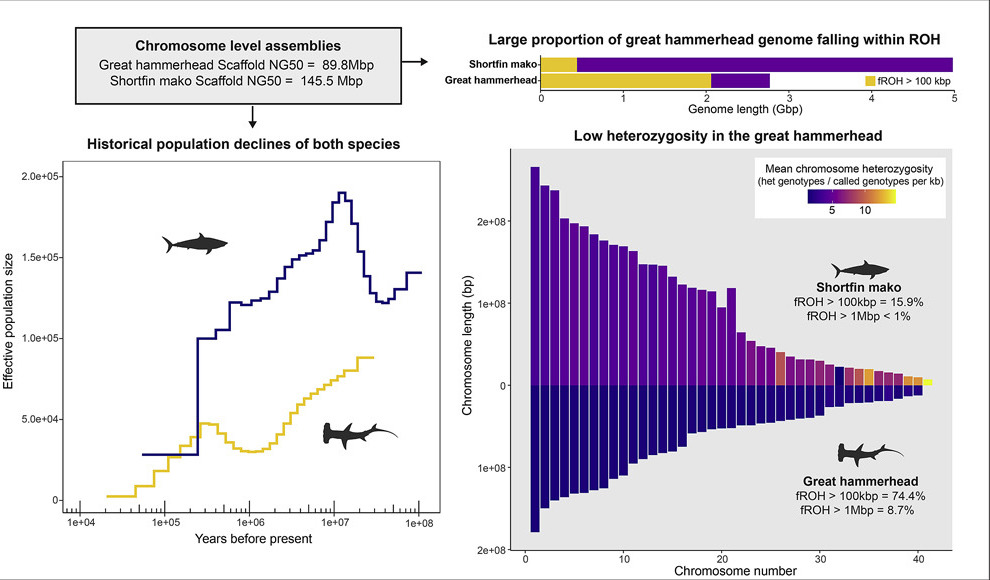
We created reference genomes for the endangered shortfin mako and critically endangered great hammerhead sharks. We detected low heterozygosity and signs of potential inbreeding in the great hammerhead, rasising concerns for the species genetic health.
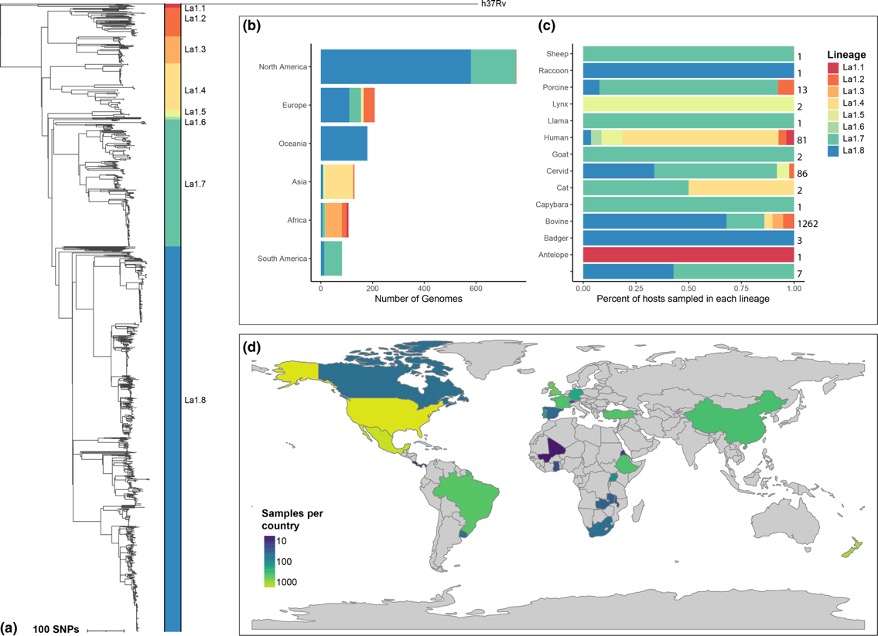
We analyzed the pangenome of M. bovis, the cause of bovine tuberculosis. We found that the accessroy genome is compact, in line with experimental studies, contradicting earlier studies which found that M. bovis has an open pangenome.
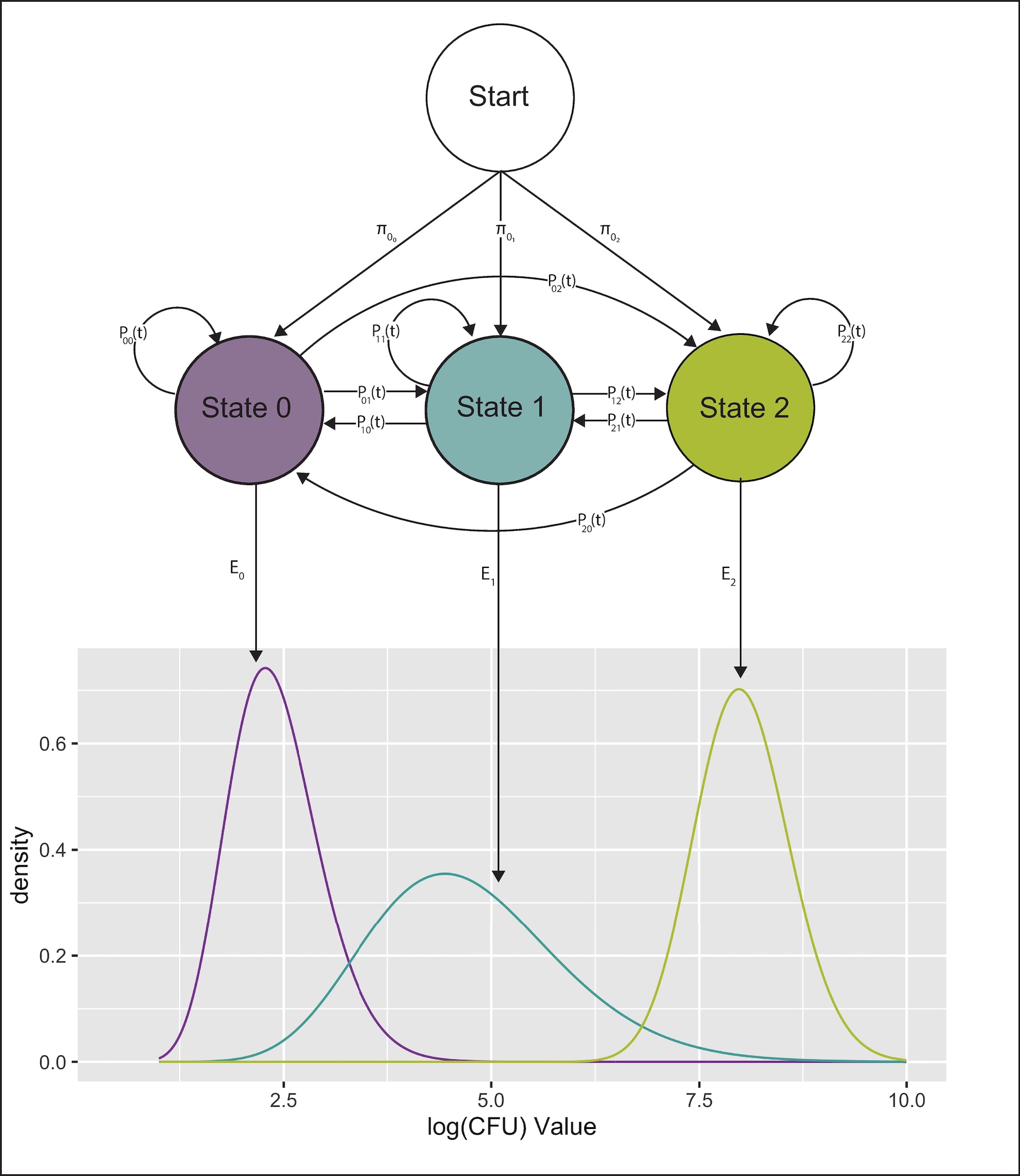
We used a continuous time HMM to characterize disease progression of Mycobacterium avium ssp. paratuberculosis infection (Johne's disease).